Data
Data
"Information, especially facts or numbers, collected to be examined and considered and used to help decision-making, or information in an electronic form that can be stored and used by a computer." (Cambridge Dictionary, 2025)
Data Challenges
Data Challenges

Data Availability
Each ML project is unique and data can be scarce
- Specific requirements
- Domain
- Location
- Space
- ...
Data Challenges
Data Usability
ML Models require data in a specific format
- Structured vs Non structured data
- Sparse data
- Legal regulations and privacy
- Storage technologies
- ...

Data Challenges
Data Quality Issues
ML Models are data-driven
- Missing values
- Duplicate records
- Incorrect data
- Outlier values
- ...

Data Challenges
Data Bias and Fairness
ML Models are data-driven
- Sampling bias
- Historical bias
- Measurement bias
- Algorithmic bias
- Implicit bias
- ...
Data Challenges
Data Complexity
Complex and dynamic systems
- Large systems
- Data generation speed
- Current systems architectures
- Interpretability issues
- Intellectual debt
- ...
The Data Science Process
The Data Science Process
The Data Science Process

The Data Science Process

Data Quality
Data Quality
Data quality refers to the state of data in terms of its fitness for a purpose

Data Quality
Data quality refers to the state of data in terms of its fitness for a purpose
This is a multi-dimensional concept
- Accuracy
- Completeness
- Uniqueness
- Consistency
- Timeliness
- Validity
Data Quality
- Data reflects reality
- Dynamic and changing data
- Confident decision-making
Accuracy
Data Quality
Metrics
- Error rate
- Precision
- Recall
- F1 score
Accuracy
Data Quality
- All the data is available
- Critical data vs Optional data
- Impact of the missing data
Completeness
Data Quality
Metrics
- Missing value ratio
- Record completeness
- Attribute completeness
Completeness
Data Quality
- Duplicated records
- Combining datasets
- Trusted data
Uniqueness
Data Quality
Metrics
- Duplicate ratio
- Unique value ratio
- Entity resolution rate
Uniqueness
Data Quality
- Data values do not conflict
- Link data from multiple sources
- Data usability
Consistency
Data Quality
Metrics
- Format consistency score
- Value consistency ratio
- Cross-field consistency rate
Consistency
Data Quality
- Data available when expected
- Context-dependent
- Added data value
Timeliness
Data Quality
Metrics
- Data freshness
- Update frequency
- Processing delay
Timeliness
Data Quality
- Data conforms to a format
- Heterogeneous non-structured data
- Automation
Validity
Data Quality
Metrics
- Format compliance rate
- Schema validation score
- Business rule compliance
Validity
Data Quality

Data Quality

Poor data quality can lead to:
- Inaccurate model predictions and reduced reliability
- Biased results
- Reduced generalisability
- Increased training time
- Higher costs
- Intellectual debt
- ...
Data Quality
- Incomplete records
- Null or NaN values
- Empty fields
- Format variations
- Unit mismatches
- Naming conventions
- Extreme values
- Measurement errors
- Data entry mistakes
Data Quality
- Incomplete records
- Null or NaN values
- Empty fields
- Format variations
- Unit mismatches
- Naming conventions
- Extreme values
- Measurement errors
- Data entry mistakes
- Random variations
- Measurement errors
- Background interference
- Sampling bias
- Selection bias
- Measurement bias
- ...
Data Assess
Data Assess

Data Assess
After collecting the data (i.e., data access), we need to perform a data assessment process to understand the data, identify and mitigate data quality issues, uncover patterns, and gain insights.
Data Assess

ML Pipelines vs ML-based Systems

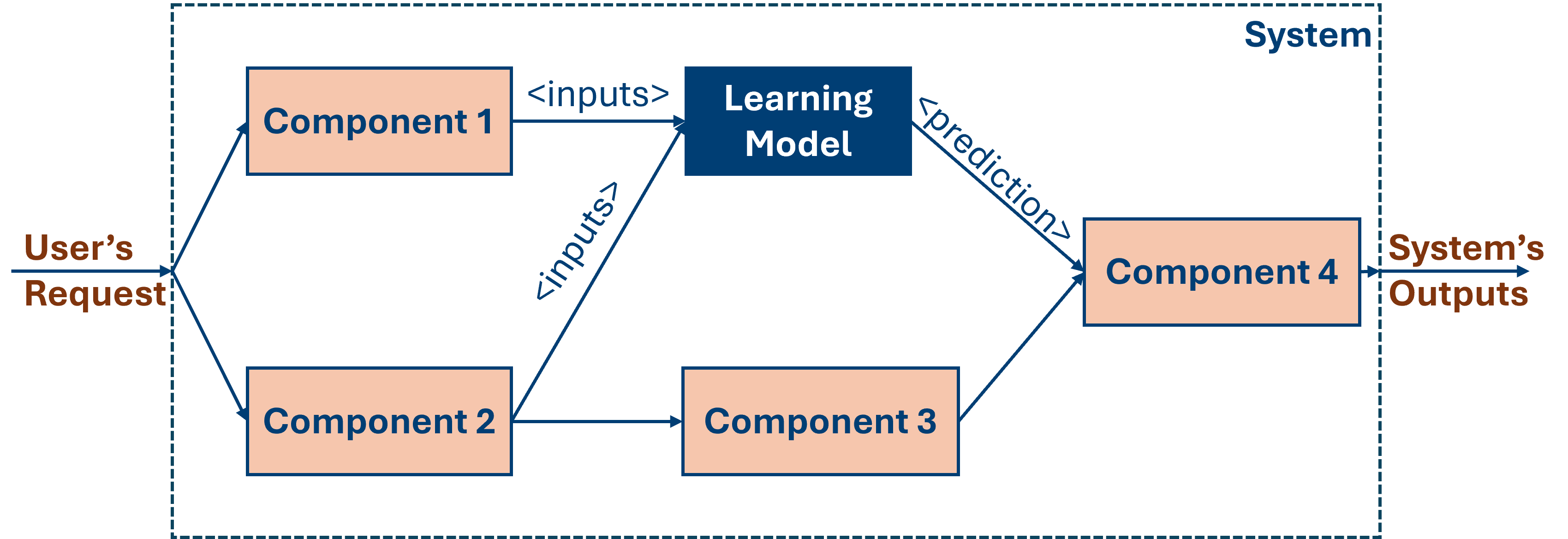
Data Assess

Data Cleaning
Process of detecting and correcting (or removing) corrupt or inaccurate records.
Data Cleaning
Missing Values
Missing data points in the dataset
- Data collection errors
- System failures
- Information not available
- Data entry mistakes
Data Cleaning
Missing Values
Missing data points in the dataset
- Data collection errors
- System failures
- Information not available
- Data entry mistakes
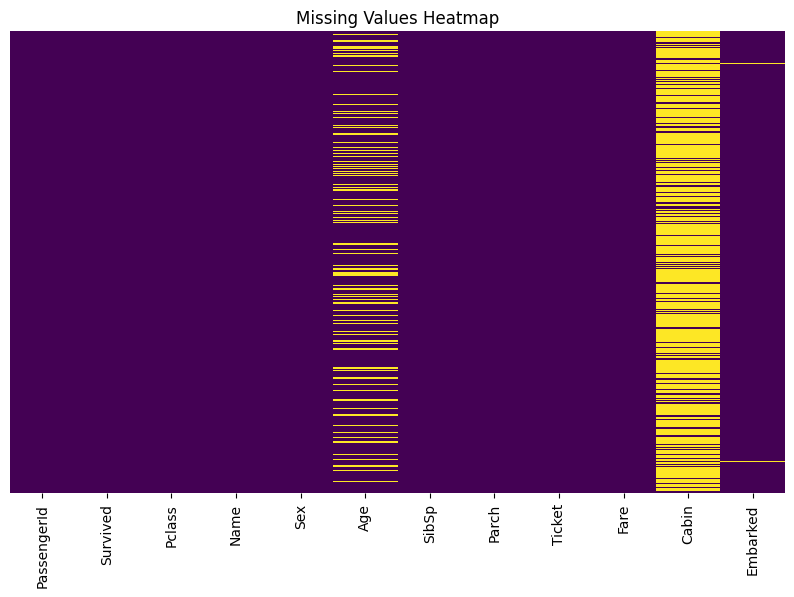
Data Cleaning
Missing Values
Missing data points in the dataset
- Deletion: Remove rows or columns with missing values
- Imputation: Fill missing values with estimated values
- Advanced techniques: Use machine learning models to predict missing values

Data Cleaning
from sklearn.linear_model import LinearRegression
features_for_age = ['Pclass', 'SibSp', 'Parch', 'Fare']
X_train = titanic_data.dropna(subset=['Age'])[features_for_age]
y_train = titanic_data.dropna(subset=['Age'])['Age']
reg_imputer = LinearRegression()
reg_imputer.fit(X_train, y_train)
X_missing = titanic_data[titanic_data['Age'].isnull()][features_for_age]
predicted_ages = reg_imputer.predict(X_missing)
titanic_data_reg = titanic_data.copy()
titanic_data_reg.loc[titanic_data_reg['Age'].isnull(), 'Age'] = predicted_ages
titanic_data['Age_Regression'] = titanic_data_reg['Age']

Data Cleaning
Outliers
Data points that significantly deviate from the rest of the data
- Measurement errors
- Data entry mistakes
- Rare but valid observations
- System malfunctions
Data Cleaning
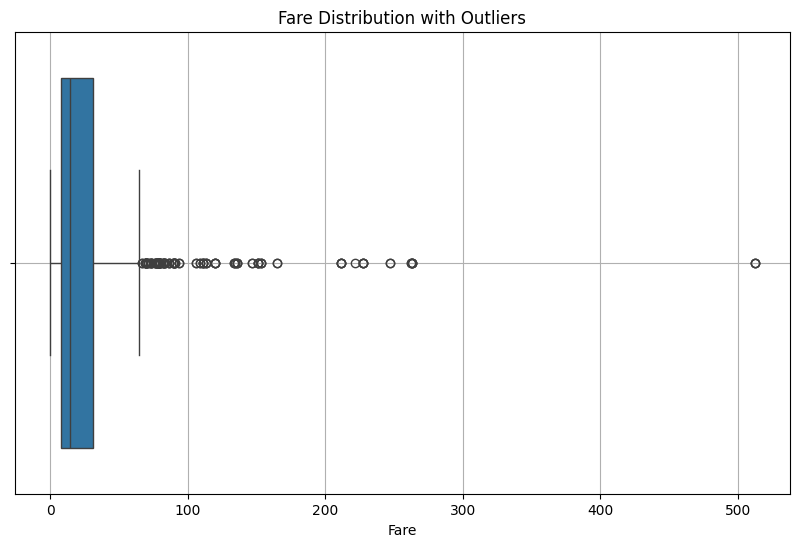
Outliers
Data points that significantly deviate from the rest of the data
- Measurement errors
- Data entry mistakes
- Rare but valid observations
- System malfunctions
Data Cleaning

Outliers
Data points that significantly deviate from the rest of the data
- Capping: Limit values to a range
- Log Transformation: Reduce the impact of extreme values
Data Cleaning

def cap_outliers(df, column):
Q1 = df[column].quantile(0.25)
Q3 = df[column].quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
df[column + '_capped'] = df[column].clip(lower=lower_bound, upper=upper_bound)
return df
Data Assess

Data Preprocessing
Data Preprocessing
Process of transforming raw data into a format suitable for machine learning while ensuring data quality and consistency.
Data Preprocessing
Feature Scaling
Transforming numerical features to a common scale
- All features contribute equally to the model
- Algorithms converge faster
- Features with larger scales do not dominate the model
Data Preprocessing
Feature Scaling
Standardisation (Z-score): Centers data around 0 with unit variance
Data Preprocessing
scaler = StandardScaler()
cols = [col + '_st' for col in num_feat]
data[cols] = scaler.fit_transform(data[num_feat])
Data Preprocessing
Feature Scaling
Min-Max scaling: Scales data to a fixed range [0,1]
Data Preprocessing
scaler = MinMaxScaler()
cols = [col + '_minmax' for col in num_feat]
data[cols] = scaler.fit_transform(data[num_feat])
Data Assess

Data Augmentation
Data Augmentation
Process of increasing the size and diversity of our datasets.
Data Augmentation
Increasing diversity to make our models more robust
- Adding controlled noise
- Random rotations
- ...
Data Augmentation
Increasing diversity to make our models more robust
- Adding controlled noise
- Random rotations
- ...
Data Augmentation
def augment_image(img, angle_range=(-15, 15)):
angle = np.random.uniform(angle_range[0], angle_range[1])
rot = rotate(img.reshape(20, 20), angle, mode='edge')
noise = np.random.normal(0, 0.05, rot.shape)
aug = rot + (noise * (rot > 0.1))
return aug.flatten()
Data Augmentation
def augment_image(img, angle_range=(-15, 15)):
angle = np.random.uniform(angle_range[0], angle_range[1])
rot = rotate(img.reshape(20, 20), angle, mode='edge')
noise = np.random.normal(0, 0.05, rot.shape)
aug = rot + (noise * (rot > 0.1))
return aug.flatten()

Data Augmentation
Increasing the size of our dataset to improve model generalisation
- Interpolating between existing data points
- Applying domain-specific transformations
- Generating synthetic data using GANs
- ...
Data Augmentation
def numerical_smote(data, k=5):
aug_data = []
for i in range(len(data)):
uniq_values = np.unique(data[data != data[i]])
dists = np.abs(uniq_values - data[i])
k_neigs = uniq_values[np.argsort(dists)[:k]]
for neig in k_neigs:
sample = data[i] + np.random.random() * (neig - data[i])
aug_data.append(sample)
return np.array(aug_data)
SMOTE (Synthetic Minority Over-sampling Technique)
- Using the k-nearest neighbours
- Interpolation between the original data point and the neighbour
Data Assess

Feature Engineering
Process of creating, transforming, and selecting features in our data, combining domain knowledge and creativity.
Feature Engineering
Creating new features in our data can help to capture important patterns or relations in the data
- Extracting information from existing features
- Combining existing features
- Adding domain knowledge rules
- ...
Feature Engineering
Creating new features in our data can help to capture important patterns or relations in the data
- Extracting information from existing features
- Combining existing features
- Adding domain knowledge rules
- ...
data['AgeGroup'] = pd.cut(data['Age'],
bins=[0, 12, 18, 35, 60],
labels=['Child',
'Teenager',
'Young Adult',
'Adult'])
Feature Engineering
Selecting the most important features in our data reduces dimensionality, prevents overfitting, improves interpretability, and reduces training time
- Correlation Matrix with Heatmap
- Decision Trees
- Principal Component Analysis (PCA)
- ...
Feature Engineering
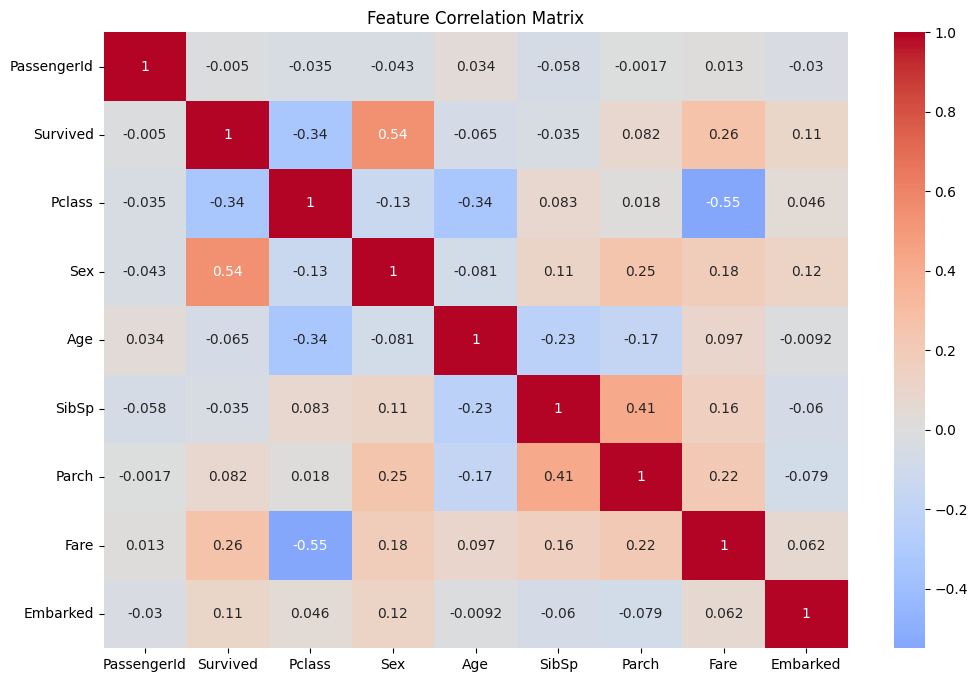
Selecting the most important features in our data reduces dimensionality, prevents overfitting, improves interpretability, and reduces training time
- Correlation Matrix with Heatmap
- Decision Trees
- Principal Component Analysis (PCA)
- ...
Feature Engineering

The matrix helps to visualise the correlation between features
Feature Engineering

corr_matrix = data.corr()
plt.figure(figsize=(12, 8))
sns.heatmap(corr_matrix, annot=True, center=0)
plt.title('Feature Correlation Matrix')
plt.show()
Feature Engineering
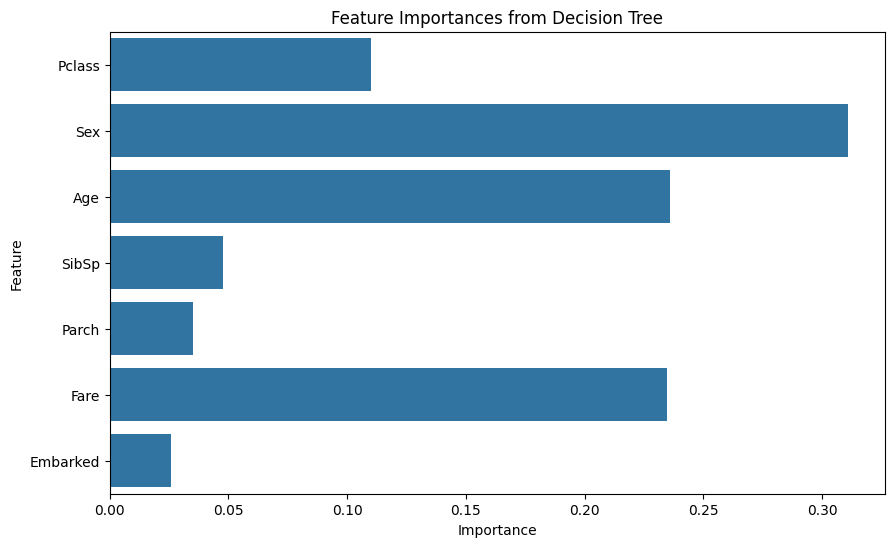
A decision tree helps in identifying the most important features contributing to the prediction. This method splits the dataset into subsets based on the feature that results in the largest information gain. At the end of the process, we get the importance of each feature
Feature Engineering

from sklearn.tree import DecisionTreeClassifier
tree = DecisionTreeClassifier(random_state=42)
tree.fit(X, y)
Feature Engineering
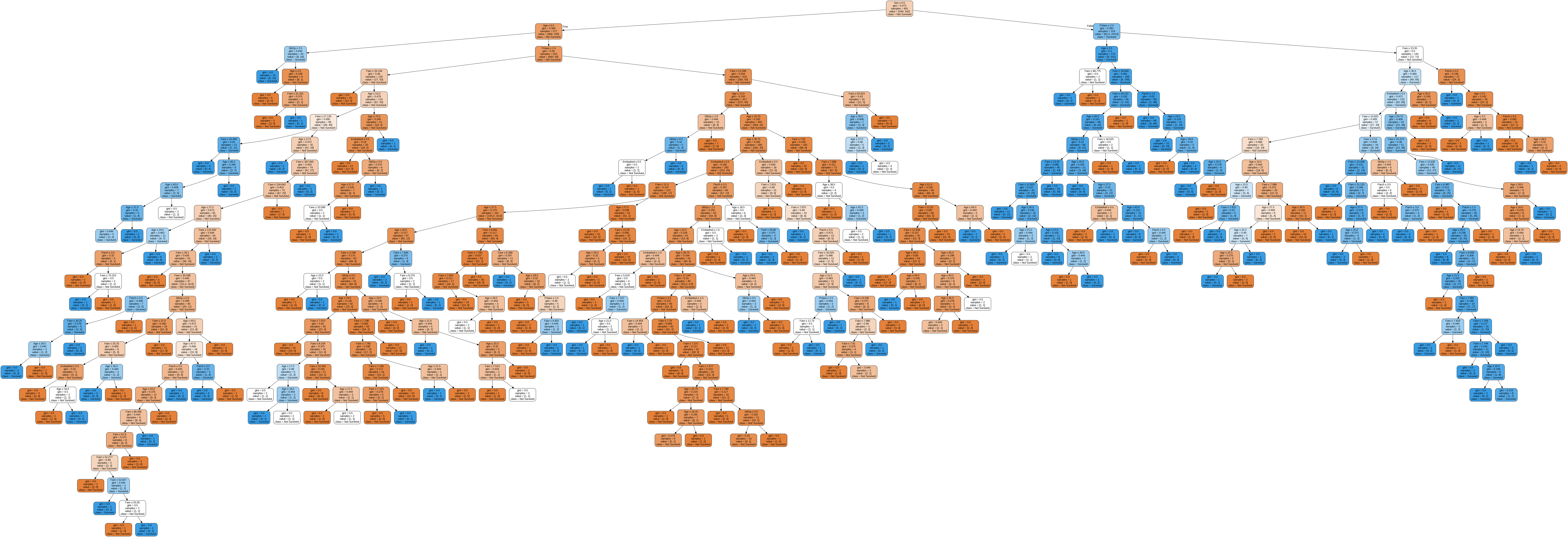
Feature Engineering
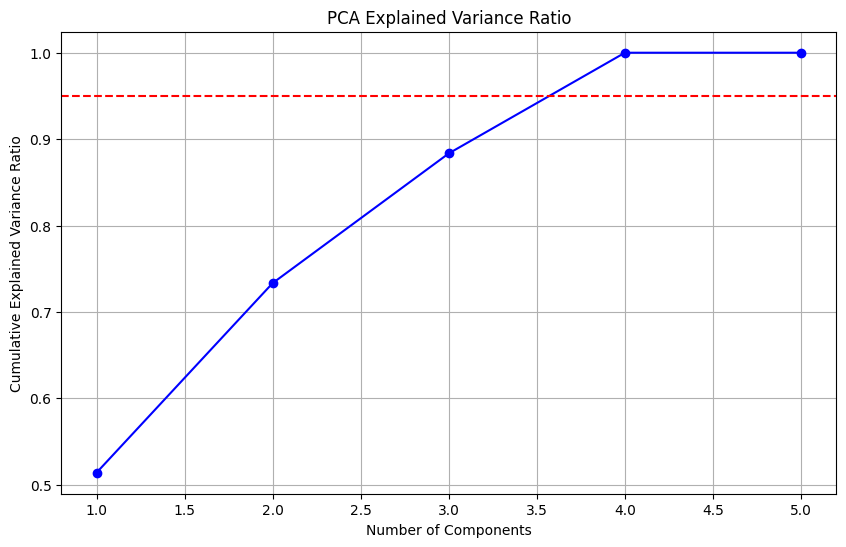
Principal Component Analysis (PCA) is a dimensionality reduction technique that transforms a set of correlated features into a set of linearly uncorrelated features called principal components. These components capture the most variance in the data, allowing for a reduction in the number of features while retaining most of the information.
Feature Engineering

from sklearn.decomposition import PCA
pca = PCA()
X_pca_transformed = pca.fit_transform(X_scaled)
explained_variance = pca.explained_variance_ratio_
cumulative_variance = np.cumsum(explained_variance)
Feature Engineering
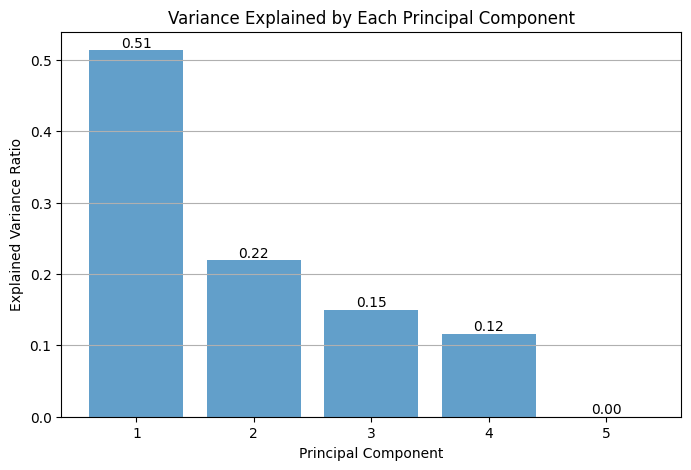
from sklearn.decomposition import PCA
pca = PCA()
X_pca_transformed = pca.fit_transform(X_scaled)
explained_variance = pca.explained_variance_ratio_
cumulative_variance = np.cumsum(explained_variance)
Data Assess

Conclusions
Overview
- Data Quality
- Data Assess
- ML Pipeline
- ML Pipeline vs ML-based Systems
- Data Cleaning and Preprocessing
- Data Augmentation
- Feature Engineering

Conclusions
Next Time
- Data Address
- Linear Regression
- Clustering

Many Thanks!
SLIDES:
_script: true
This script will only execute in HTML slides
_script: true